May 20, 2015 From rOpenSci (https://deploy-preview-304--ropensci.netlify.app/blog/2015/05/20/database-interfaces/). Except where otherwise noted, content on this site is licensed under the CC-BY license.
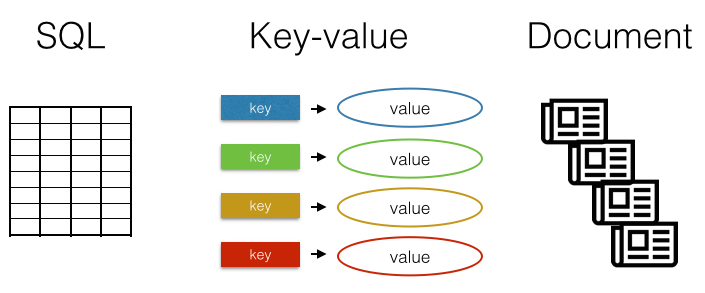
There are many different databases. The most familiar are row-column SQL databases like MySQL, SQLite, or PostgreSQL. Another type of database is the key-value store, which as a concept is very simple: you save a value specified by a key, and you can retrieve a value by its key. One more type is the document database, which instead of storing rows and columns, stores blobs of text or even binary files. The key-value and document types fall under the NoSQL umbrella. As there are mature R clients for many SQL databases, and dplyr is a great generic interface to SQL backends (see dplyr vignettes for an intro), we won’t delve into SQL clients here.
What is the difference between SQL and NoSQL (key-value, document)? A diagram may be helpful:
gistr map
NoSQL is often interpreted as Not only SQL - meaning a database that is called a NoSQL database may contain some notion of row-column storage, but other details diverge from traditional SQL databases. See Wikipedia for more information.
If you aren’t already using databases, why care about databases? We’ll answer this through a number of use cases:
- Use case 1: Let’s say you are producing a lot of data in your lab - millions of rows of data. Storing all this data in
.xlsor.csvfiles would definitely get cumbersome. If the data is traditional row-column spreadsheet data, a SQL database is perfect, perhaps PostgreSQL. Putting your data in a database allows the data to scale up easily while maintaining speedy data access, many tables can be linked together if needed, and more. Of course if your data will always fit in memory of the machine you’re working on, a database may be too much complexity. - Use case 2: You already have data in a database, whether SQL or NoSQL. Of course it makes sense to interface with the database from R instead of e.g., exporting files from the database, then into R.
- Use case 3: A data provider gives dumps of data that you need for your research/work problem. You download the data and it’s hundreds of
.csvfiles. It sure would be nice to be able to efficiently search this data. Simple searches like return all records with variable X < 10 are ideal for a SQL database. If instead the files are blobs of XML, JSON, or something else non-tabular, a document database is ideal. - Use case 4: You need to perform more complicated searches than SQL can support. Some NoSQL databases have very powerful search engines, e.g., Elasticsearch.
- Use case 5: Sometimes you just need to cache stuff. Caching is a good use case for key-value stores. Let’s say you are requesting data from a database online, and you want to make a derivative data thing from the original data, but you don’t want to lose the original data. Simply storing the original data on disk in files is easy and does the job. Sometimes though, you may need something more structured. Redis and etcd are two key-value stores we make clients for and can make caching easy. Another use for caching is to avoid repeating time-consuming queries or queries that may cost money.
- Use case 6: Indexable serialization. Related to the previous discussion of caching, this is caching, but better. That is, instead of dumping an R object to a cache, then retrieving the entire object later, NoSQL DB’s make it easy to serialize an R object, and retrieve only what you need. See Rich FitzJohn’s storr for an example of this.
rOpenSci has an increasing suite of database tools:
- elastic (document database) (on CRAN)
- sofa (document database)
- solr (document database) (on CRAN)
- etseed (key-value store)
- rrlite (key-value store)
- rerddap (SQL database as a service, open source) (on CRAN)
- ckanr (SQL database as a service, open source)
- nodbi (DBI, but for NoSQL DB’s)
Some of these packages (e.g., rrlite, nodbi) can be thought of as infrastructure, just like clients for PostgreSQL or SQLite, for which other R packages can be created - or that can be used to interface with a database. Other packages (e.g., ckanr) are more likely to be useful to end users for retrieving data for a project.
If you’re wondering what database to use:
- You may want a SQL database if: you have tabular data, and the schema is not going to change
- You may want a NoSQL key-value database if: you want to shove objects into something, and then retrieve later by a key
- You may want a NoSQL document database if:
- you need to store unstructured blobs, even including binary attachments
- you need a richer query interface than a SQL database can provide
SQL databases have many advantages - one important advantage is that SQL syntax is widely used, and there are probably clients in every concievable language for interacting with SQL databases. However, NoSQL can be a better fit in many cases, overriding this SQL syntax familiarity.
There is another type of NoSQL database, the graph database, including Neo4j and Titan. We didn’t talk much about them here, but they can be useful when you have naturally graph like data. A science project using a graph database is Open Tree of Life. There is an R client for Neo4J: RNeo4j.
Let us know if you have any feedback on these packages, and/or if you think there’s anything else we should be thinking about making in this space. Now on to some examples of rOpenSci packages.
🔗 Get devtools
We’ll need devtools to install some of these packages, as not all are on CRAN. If you are on Windows, see these notes.
install.packages("devtools")
🔗 elastic
elastic - Interact with Elasticsearch.
install.packages("elastic")
library("elastic")
elastic is a powerful document database with a built in query engine. It speaks JSON, has a nice HTTP API, which we use to communicate with elastic from R. What’s great about elastic over e.g., Solr is that you don’t have to worry about specifying a schema for your data. You can simply put data in, and then query on that data. You can specify configuration settings.
🔗 Example
In a quick example, here’s going from a data.frame in R, putting data into elastic, then querying on that data.
library("ggplot2")
invisible(connect())
res <- docs_bulk(diamonds, "diam")
About 54K records in Elasticsearch for the dataset.
count("diam")
#> [1] 53940
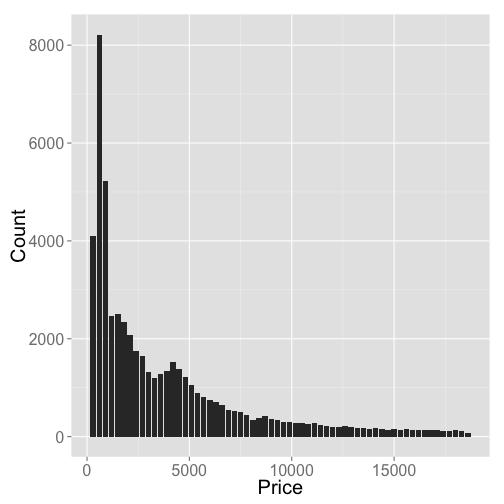
We don’t have time to go through hardly any of the diverse and powerful Elasticsearch query interface, so as an example, let’s plot the price of diamonds in $300 buckets using the Elasticsearch aggregations search API
aggs <- '{
"aggs": {
"pricebuckets" : {
"histogram" : {
"field" : "price",
"interval" : 300
}
}
}
}'
res <- Search("diam", body = aggs, size = 0)
df <- do.call("rbind.data.frame", res$aggregations$pricebuckets$buckets)
ggplot(df, aes(key, doc_count)) +
geom_bar(stat = "identity") +
theme_grey(base_size = 20) +
labs(x = "Price", y = "Count")
gistr map
We have a package in developmented called elasticdsl that follows the lead of the Python client elasticsearch-dsl-py to allow native R based ways to specify queries. The package focuses on querying for data, whereas other operations will remain in the lower level elastic client.
🔗 sofa
sofa - Interact with CouchDB.
devtools::install_github("ropensci/sofa")
library("sofa")
🔗 Example
Connect to your running CouchDB instance:
?cushion
Create a database
db_create(dbname = 'sofadb')
Create a document in that database
doc_create('{"name":"sofa","beer":"IPA"}', dbname = "sofadb", docid = "a_beer")
Get the document
doc_get(dbname = "sofadb", docid = "a_beer")
There’s a similar interface to inserting data within R directly into CouchDB, just as with Elasticsearch above. There’s lots more to do in sofa, including adding ability to do map-reduce.
🔗 solr
solr - A client for interacting with Solr.
solr focuses on reading data from Solr engines. We are working on adding functionality for working with more Solr features, including writing documents. Adding support for writing to solr is a bit trickier than reading data, since writing data requires specifying a schema.
install.packages("solr")
library("solr")
🔗 Example
A quick example using a remote Solr server the Public Library of Science search engine.
solr_search(q = '*:*', fl = c('id', 'journal', 'publication_date'), base = 'http://api.plos.org/search', verbose = FALSE)
#> id journal
#> 1 10.1371/journal.pone.0123754/title PLOS ONE
#> 2 10.1371/journal.pone.0123754/abstract PLOS ONE
#> 3 10.1371/journal.pone.0123754/references PLOS ONE
#> 4 10.1371/journal.pone.0123754/body PLOS ONE
#> 5 10.1371/journal.pone.0123754/introduction PLOS ONE
#> 6 10.1371/journal.pone.0123754/results_and_discussion PLOS ONE
#> 7 10.1371/journal.pone.0123754/materials_and_methods PLOS ONE
#> 8 10.1371/journal.pone.0123754/conclusions PLOS ONE
#> 9 10.1371/journal.pone.0031384 PLoS ONE
#> 10 10.1371/journal.pone.0031384/title PLoS ONE
#> publication_date
#> 1 2015-05-04T00:00:00Z
#> 2 2015-05-04T00:00:00Z
#> 3 2015-05-04T00:00:00Z
#> 4 2015-05-04T00:00:00Z
#> 5 2015-05-04T00:00:00Z
#> 6 2015-05-04T00:00:00Z
#> 7 2015-05-04T00:00:00Z
#> 8 2015-05-04T00:00:00Z
#> 9 2012-02-14T00:00:00Z
#> 10 2012-02-14T00:00:00Z
solr is quite useful in R since it is a common search engine that is often exposed as is, so that you can pop this solr R client into your script or package and (hopefully) not have to worry about how to query the Solr service.
🔗 etseed
etseed is an R client for the etcd key-value store, developed by the folks at coreos, written in Go.
This package is still in early days, and isn’t exactly the fastest option in the bunch here - but upcoming changes (including allowing bulk writing and retrieval) in etcd should help.
devtools::install_github("ropensci/etseed")
library("etseed")
A note before we go through an example. etcd has a particular way of specifying keys, in that you have to prefix a key by a forward slash, like
/foobarinstead offoobar.
🔗 Example
Save a value to a key
create(key = "/mykey", value = "this is awesome")
#> $action
#> [1] "set"
#>
#> $node
#> $node$key
#> [1] "/mykey"
#>
#> $node$value
#> [1] "this is awesome"
#>
#> $node$modifiedIndex
#> [1] 1299
#>
#> $node$createdIndex
#> [1] 1299
#>
#>
#> $prevNode
#> $prevNode$key
#> [1] "/mykey"
#>
#> $prevNode$value
#> [1] "this is awesome"
#>
#> $prevNode$modifiedIndex
#> [1] 1298
#>
#> $prevNode$createdIndex
#> [1] 1298
Fetch the value given a key
key(key = "/mykey")
#> $action
#> [1] "get"
#>
#> $node
#> $node$key
#> [1] "/mykey"
#>
#> $node$value
#> [1] "this is awesome"
#>
#> $node$modifiedIndex
#> [1] 1299
#>
#> $node$createdIndex
#> [1] 1299
🔗 rrlite
rrlite - An R client for the Redis C library rlite
devtools::install_github("ropensci/rrlite")
library("rrlite")
This package may be more interesting than other R Redis clients because there is no need to start up a server since rlite is a serverless engine.
🔗 Example
Here, we initialize, then put 20 values into rlite, assigned to the key foo, then retrieve the values by the same key.
r <- RedisAPI::rdb(rrlite::hirlite)
r$set("foo", runif(20))
r$get("foo")
#> [1] 0.51670270 0.08039860 0.34762872 0.30276370 0.15985876 0.66062207
#> [7] 0.26802708 0.97451274 0.94458185 0.04604044 0.93153133 0.91241321
#> [13] 0.64395377 0.12517230 0.31826622 0.34425757 0.79364064 0.91926051
#> [19] 0.47487029 0.11644076
This is a good candidate for using within other R packages for more sophisticated caching than simply writing to disk, and is especially easy since users aren’t required to spin up a server as with normal Redis, or other DB’s like CouchDB, MongoDB, etc.
🔗 rerddap
rerddap - A general purpose R client for any ERDDAP server.
ERDDAP servers
install.packages("rerddap")
library("rerddap")
ERDDAP is a server built on top of OPenDAP. NOAA serve many differen datasets through ERDDAP servers. Through ERDDAP, you can get gridded data (see griddap()), which lets you query from gridded datasets (see griddap()), or tablular datasets (see tabledap()). ERDDAP is open source, so anyone can use it to serve data.
rerddap by default grabs NetCDF files, a binary compressed file type that should be faster to download, and take up less disk space, than other formats (e.g., csv). However, this means that you need a client library for NetCDF files - but not to worry, we use ncdf by default (for which there are CRAN binaries for all platforms), but you can choose to use ncdf4 (binaries only for some platforms).
🔗 Example
In this example, we search for gridded datasets
ed_search(query = 'size', which = "grid")
#> 6 results, showing first 20
#> title dataset_id
#> 11 NOAA Global Coral Bleaching Monitoring Products NOAA_DHW
#> 13 USGS COAWST Forecast, US East Coast and Gulf of Mexico (Experimental) [time][s_rho][eta_rho][xi_rho] whoi_7dd7_db97_4bbe
#> 14 USGS COAWST Forecast, US East Coast and Gulf of Mexico (Experimental) [time][Nbed][eta_rho][xi_rho] whoi_a4fb_2c9c_16a7
#> 15 USGS COAWST Forecast, US East Coast and Gulf of Mexico (Experimental) [time][eta_rho][xi_rho] whoi_ed12_89ce_9592
#> 16 USGS COAWST Forecast, US East Coast and Gulf of Mexico (Experimental) [time][eta_u][xi_u] whoi_61c3_0b5d_cd61
#> 17 USGS COAWST Forecast, US East Coast and Gulf of Mexico (Experimental) [time][eta_v][xi_v] whoi_62d0_9d64_c8ff
Get more information on a single dataset of interest
info('noaa_esrl_027d_0fb5_5d38')
#> <ERDDAP info> noaa_esrl_027d_0fb5_5d38
#> Dimensions (range):
#> time: (1850-01-01T00:00:00Z, 2014-05-01T00:00:00Z)
#> latitude: (87.5, -87.5)
#> longitude: (-177.5, 177.5)
#> Variables:
#> air:
#> Range: -20.9, 19.5
#> Units: degC
Then fetch the dataset
griddap('noaa_esrl_027d_0fb5_5d38',
time = c('2012-07-01', '2012-09-01'),
latitude = c(21, 19),
longitude = c(-80, -76)
)
#> <ERDDAP griddap> noaa_esrl_027d_0fb5_5d38
#> Path: [~/.rerddap/3d23f817218694a622895e78417d291f.nc]
#> Last updated: [2015-05-14 11:02:25]
#> File size: [0 mb]
#> Dimensions (dims/vars): [3 X 1]
#> Dim names: time, latitude, longitude
#> Variable names: CRUTEM3: Surface Air Temperature Monthly Anomaly
#> data.frame (rows/columns): [6 X 4]
#> time latitude longitude air
#> 1 2012-07-01T00:00:00Z 22.5 -77.5 NA
#> 2 2012-07-01T00:00:00Z 22.5 -77.5 0.2500000
#> 3 2012-08-01T00:00:00Z 22.5 -77.5 NA
#> 4 2012-08-01T00:00:00Z 17.5 -77.5 0.2666667
#> 5 2012-09-01T00:00:00Z 17.5 -77.5 NA
#> 6 2012-09-01T00:00:00Z 17.5 -77.5 0.1000000
There are many different ERDDAP servers, see the function servers() for help.
🔗 ckanr
ckanr - A general purpose R client for any CKAN server.
CKAN is similar to ERDDAP in being an open source system to store and provide data via web services (and web interface, but we don’t need that here). CKAN bills itself as an open-source data portal platform.
devtools::install_github("ropensci/ckanr")
library("ckanr")
🔗 Example
Examples use the CKAN server at http://data.techno-science.ca
Show changes in a CKAN server
changes(limit = 10, as = "table")[, 1:2]
#> user_id timestamp
#> 1 27778230-2e90-4818-9f00-bbf778c8fa09 2015-03-30T15:06:55.500589
#> 2 27778230-2e90-4818-9f00-bbf778c8fa09 2015-01-09T23:33:14.303237
#> 3 27778230-2e90-4818-9f00-bbf778c8fa09 2015-01-09T23:31:49.537792
#> 4 b50449ea-1dcc-4d52-b620-fc95bf56034b 2014-11-06T18:58:08.001743
#> 5 b50449ea-1dcc-4d52-b620-fc95bf56034b 2014-11-06T18:55:55.059527
#> 6 27778230-2e90-4818-9f00-bbf778c8fa09 2014-11-05T23:17:46.422404
#> 7 27778230-2e90-4818-9f00-bbf778c8fa09 2014-11-05T23:17:05.134909
#> 8 27778230-2e90-4818-9f00-bbf778c8fa09 2014-11-05T23:12:44.074493
#> 9 27778230-2e90-4818-9f00-bbf778c8fa09 2014-11-05T23:11:41.536040
#> 10 27778230-2e90-4818-9f00-bbf778c8fa09 2014-11-05T21:54:39.496994
Search for data packages
package_search(q = '*:*', rows = 2, as = "table")$results[, 1:7]
#> license_title maintainer relationships_as_object private maintainer_email revision_timestamp
#> 1 Open Government Licence - Canada NULL FALSE 2014-10-28T21:18:27.068320
#> 2 Open Government Licence - Canada NULL FALSE 2014-10-28T21:18:58.958555
#> id
#> 1 f4406699-3e11-4856-be48-b55da98b3c14
#> 2 0a801729-aa94-4d76-a5e0-7b487303f4e5
More information on CKAN: https://docs.ckan.org/en/latest/contents.html
🔗 nodbi
nodbi - Like the DBI package, but for document and key-value databases.
nodbi has five backends at the moment:
- Redis
- etcd
- MongoDB
- CouchDB
- Elasticsearch
nodbi is in early development, so expect changes - but that also means it’s a good time to give your input. What use cases you can think of for this package? What database do you think should be added as a backend?
devtools::install_github("ropensci/nodbi")
library("nodbi")
🔗 Example
We’ll use MongoDB to store some data, then pull it back out. First, start up your mongo server, then intialize the connection
mongod
(src <- src_mongo())
#> MongoDB 3.0.2 (uptime: 230s)
#> URL: Scotts-MBP/test
Insert data
library("ggplot2")
diamonds$cut <- as.character(diamonds$cut)
diamonds$color <- as.character(diamonds$color)
diamonds$clarity <- as.character(diamonds$clarity)
docdb_create(src, key = "diam", value = diamonds)
Pull data back out
res <- docdb_get(src, "diam")
head(res)
#> carat cut color clarity depth table price x y z
#> 1 0.23 Ideal E SI2 61.5 55 326 3.95 3.98 2.43
#> 2 0.21 Premium E SI1 59.8 61 326 3.89 3.84 2.31
#> 3 0.23 Good E VS1 56.9 65 327 4.05 4.07 2.31
#> 4 0.29 Premium I VS2 62.4 58 334 4.20 4.23 2.63
#> 5 0.31 Good J SI2 63.3 58 335 4.34 4.35 2.75
#> 6 0.24 Very Good J VVS2 62.8 57 336 3.94 3.96 2.48
Data is identical:
identical(diamonds, res)
#> [1] TRUE

