July 16, 2019 From rOpenSci (https://deploy-preview-304--ropensci.netlify.app/blog/2019/07/16/grainchanger/). Except where otherwise noted, content on this site is licensed under the CC-BY license.
The grainchanger package provides functionality for data aggregation to a coarser resolution via moving-window or direct methods.
🔗 Why do we need new methods for data aggregation?
As landscape ecologists and macroecologists, we often need to aggregate data in order to harmonise datasets. In doing so, we often lose a lot of information about the spatial structure and environmental heterogeneity of data measured at finer resolution.
The issues around scale disconnects are both conceptual and practical:
🔗 Conceptual issues
-
The scale at which drivers affect responses varies
-
As does the resolution at which broad-scale patterns of ecological processes emerge
🔗 Technical issues
-
The scale we measure this stuff at varies too
-
Ecological response data (e.g. species richness from species' atlas data) are often available at coarse resolutions, whereas with increasing availability of high-resolution remote sensing data, environmental data are available at much finer resolutions
So how do we aggregate this fine-resolution data without losing information about spatial structure? We argue that using a moving window to calculate heterogeneity before aggregating retains information about spatial structure at an appropriate scale. We can then use this aggregated measure to (1) identify the appropriate scale-of-effect when we only have coarse-grain response data; and (2) investigate the effect of cross-scale interactions on ecological phenomena. See our paper in Methods in Ecology & Evolution1 for simulations and a case study which show this.
We developed this method and R package in order to overcome some of these issues.
🔗 How does the method work?
The moving-window data aggregation (MWDA) method smooths an input raster using a specified function within a moving window of a specified size and shape prior to aggregation.
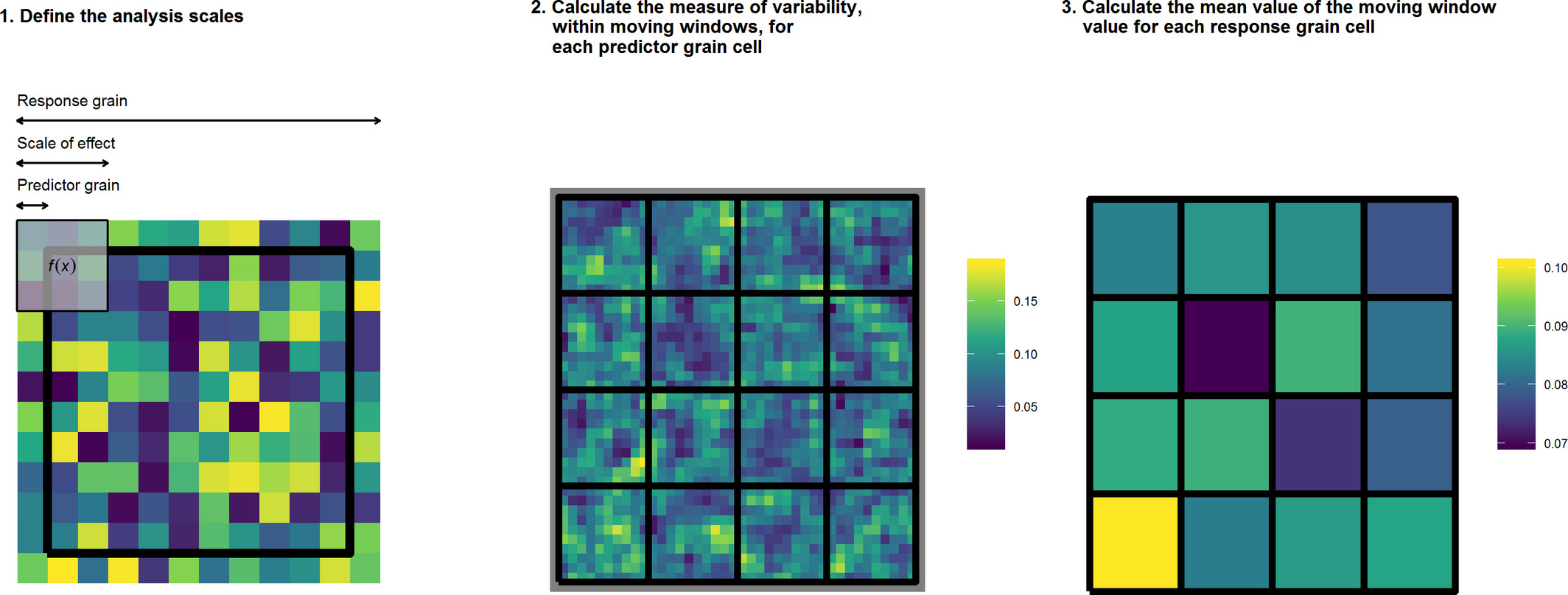
The above is a graphical representation of the MWDA method. In calculating the MWDA measure, three aspects of scale are considered. Predictor grain is the characteristic spatial scale of a predictor variable, that is, the resolution of the environmental data; scale‐of‐effect determines the appropriate scale of the relationship between predictor and response, for example, an ecological neighbourhood; response grain is the grain of the unit into which you are predicting, that is, the resolution of the response variable (represented by the black lines). Note that the colour scale is unitless. Yellow cells represent ‘high’ values and dark blue cells ‘low’ values. Panel 1 shows a close up of one of the response grain cells in panel 2, whereas panel 2 shows all response grain cells for the study region. Panel 3 shows the study region after aggregation.
The package rests around three key functions:
winmovewinmove_aggnomove_agg
In winmove the user can smooth a raster using a window of a specified shape and size and a specified function (either those that are built in, or a user defined function).
In winmove_agg, the user specifies an aggregation function in addition to the above.
In nomove_agg, the user specifies only the aggregation function.
Below is an example of the three functions applied to a continuous landscape with the moving window function as var within a square window of 5 units. For winmove_agg the aggregation function is mean, for nomove_agg it is var.
library(grainchanger)
library(ggplot2)
library(landscapetools)
library(patchwork)
# continuous landscape
p1 <- show_landscape(cont_ls,
legend_title = "",
xlab = "", ylab = "") +
ggtitle("Landscape") +
theme_bw() +
coord_equal()
# moving-window smoothing using variance
win_smooth <- winmove(fine_dat = cont_ls,
d = 5,
type = "rectangle",
win_fun = var)
p2 <- show_landscape(win_smooth,
legend_title = "",
xlab = "", ylab = "") +
ggtitle("winmove") +
theme_bw() +
coord_equal()
# moving-window aggregation using variance
g_sf$mwda <- winmove_agg(coarse_dat = g_sf,
fine_dat = cont_ls,
d = 5,
type = "rectangle",
win_fun = var,
agg_fun = mean,
quiet = TRUE)
p3 <- ggplot(g_sf) +
geom_sf(aes(fill = mwda)) +
scale_fill_viridis_c(name = "") +
theme_bw() +
ggtitle("winmove_agg")
# standard aggregation using variance
g_sf$dda <- nomove_agg(coarse_dat = g_sf,
fine_dat = cont_ls,
agg_fun = var,
quiet = TRUE)
p4 <- ggplot(g_sf) +
geom_sf(aes(fill = dda)) +
scale_fill_viridis_c(name = "") +
theme_bw() +
ggtitle("nomove_agg")
(p1 + p2) / (p3 + p4)

See the package website for a description of the built-in functions.
🔗
Why not just use the raster package?
The key functionality of grainchanger is moving window smoothing, and aggregation to a coarser resolution. “But aren’t these available in the raster package?”, I hear you ask.
While raster::focal is super fast for calculating using sum, on large rasters it can become computationally cumbersome with other functions. However, using the weights matrix and the sum function, it is possible to calculate many other functions. Our package provides a wrapper to do so.
As a side note, it was a helpful post from whuber on StackExchange which led me to realise this.
Additionally, the aggregation functions in the grainchanger package have been written so that (a) the user can aggregate to non-rectangular shapes; and (b) it can be run in parallel using the future package. This is particularly advantageous when using winmove_agg. Parallelisation can be achieved by adding the following line prior to running either winmove_agg or nomove_agg:
plan(multiprocess)
🔗 Other features
In testing the method, we needed to simulate landscapes (which we did with a great rOpenSci package NLMR). To avoid edge effects, we wanted to create the effect of a torus (an infinite surface where the top joins the bottom, and the left side meets the right side), but could not find a function to do so - so we wrote one instead. Hopefully others will find this useful when working with simulated landscapes.
torus <- create_torus(cat_ls, 15)
p1 <- show_landscape(cat_ls,
legend_title = "",
xlab = "", ylab = "") +
ggtitle("Landscape")
p2 <- show_landscape(torus,
legend_title = "",
xlab = "", ylab = "") +
ggtitle("Landscape + torus")
p1 + p2

🔗 Contributing
We hope to continue to build on the functions so that they are as optimised as we can make them. If you have a particular function you would like to see included in the package, please create a new issue, or submit a pull request making sure develop is the destination branch on the grainchanger repository.
In the long term, we would like to move the package into C++ so that all functions are fast. If you are interested in collaborating on this please get in touch.
🔗 Acknowledgements
I would really like to acknowledge the rOpenSci review process. The open nature of the reviews made for a thoughtful process, and I learned a lot from it. Thanks to Max Joseph and John Baumgartner for their time and expertise. The package and it’s documentation are much improved thanks to their input. Felix Eigenbrod and I worked on the methods behind the package together as part of the ERC funded project Scaling rules for ecosystem services. Thanks also for Marco Sciaini for helpful discussion around how to optimise some of this code and the tips on R package development.

